2020¶
Estimating the effective reproduction number in Belgium with the RKI method
Using the Robert Koch Institute method with serial interval of 4.
Every day Bart Mesuere tweets a nice dashboard with current numbers about Covid-19 in Belgium. This was the tweet on Wednesday 20/11/04:
twitter: https://twitter.com/BartMesuere/status/1323881489864548352
It's nice to see that the effective reproduction number ($Re(t)$) is again below one. That means the power of virus is declining and the number of infection will start to lower. This occured first on Tuesday 2020/11/3:
twitter: https://twitter.com/BartMesuere/status/1323519613855059968
I estimated the $Re(t)$ earlier with rt.live model in this notebook. There the $Re(t)$ was still estimated to be above one. Michael Osthege replied with a simulation results with furter improved model:
twitter: https://twitter.com/theCake/status/1323211910481874944
In that estimation, the $Re(t)$ was also not yet heading below one at the end of october.
In this notebook, we will implement a calculation based on the method of the Robert Koch Institute. The method is described and programmed in R in this blog post.
In that blogpost there's a link to a website with estimations for most places in the world The estimation for Belgium is here
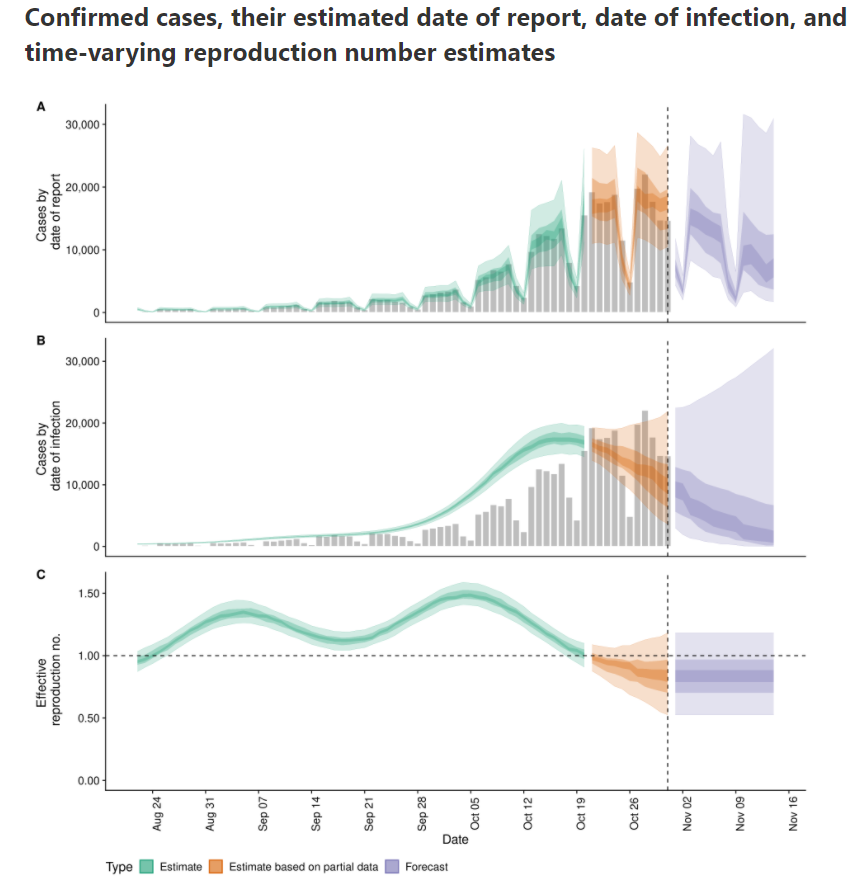
According to that calculation, $Re(t)$ is already below zero for some days.
Load libraries and data
df_tests = pd.read_csv('https://epistat.sciensano.be/Data/COVID19BE_tests.csv', parse_dates=['DATE'])
df_cases = pd.read_csv('https://epistat.sciensano.be/Data/COVID19BE_CASES_AGESEX.csv', parse_dates=['DATE'])
df_cases
| DATE | PROVINCE | REGION | AGEGROUP | SEX | CASES | |
|---|---|---|---|---|---|---|
| 0 | 2020-03-01 | Antwerpen | Flanders | 40-49 | M | 1 |
| 1 | 2020-03-01 | Brussels | Brussels | 10-19 | F | 1 |
| 2 | 2020-03-01 | Brussels | Brussels | 10-19 | M | 1 |
| 3 | 2020-03-01 | Brussels | Brussels | 20-29 | M | 1 |
| 4 | 2020-03-01 | Brussels | Brussels | 30-39 | F | 1 |
| ... | ... | ... | ... | ... | ... | ... |
| 36279 | NaT | VlaamsBrabant | Flanders | 40-49 | M | 3 |
| 36280 | NaT | VlaamsBrabant | Flanders | 50-59 | M | 1 |
| 36281 | NaT | WestVlaanderen | Flanders | 20-29 | F | 1 |
| 36282 | NaT | WestVlaanderen | Flanders | 50-59 | M | 3 |
| 36283 | NaT | NaN | NaN | NaN | NaN | 1 |
36284 rows × 6 columns
Reformat data into Rtlive format
df_cases_per_day = (df_cases
.dropna(subset=['DATE'])
.assign(region='Belgium')
.groupby(['region', 'DATE'], as_index=False)
.agg(cases=('CASES', 'sum'))
.rename(columns={'DATE':'date'})
.set_index(["region", "date"])
)
What's in our basetable:
| cases | ||
|---|---|---|
| region | date | |
| Belgium | 2020-03-01 | 19 |
| 2020-03-02 | 19 | |
| 2020-03-03 | 34 | |
| 2020-03-04 | 53 | |
| 2020-03-05 | 81 | |
| ... | ... | |
| 2020-11-01 | 2660 | |
| 2020-11-02 | 13345 | |
| 2020-11-03 | 11167 | |
| 2020-11-04 | 4019 | |
| 2020-11-05 | 5 |
250 rows × 1 columns
Let's plot the number of cases in function of the time.
ax = df_cases_per_day.loc['Belgium'].plot(figsize=(18,6))
ax.set(ylabel='Number of cases', title='Number of cases for covid-19 and number of positives in Belgium');

We see that the last days are not yet complete. Let's cut off the last two days of reporting.
Calculate the date two days ago:
datetime.date(2020, 11, 3)
# today_minus_two = datetime.date.today() + relativedelta(days=-2)
today_minus_two = datetime.date(2020, 11, 3) # Fix the day
today_minus_two.strftime("%Y-%m-%d")
'2020-11-03'
Replot the cases:
ax = df_cases_per_day.loc['Belgium'][:today_minus_two].plot(figsize=(18,6))
ax.set(ylabel='Number of cases', title='Number of cases for covid-19 and number of positives in Belgium');

Select the Belgium region:
| cases | |
|---|---|
| date | |
| 2020-03-01 | 19 |
| 2020-03-02 | 19 |
| 2020-03-03 | 34 |
| 2020-03-04 | 53 |
| 2020-03-05 | 81 |
| ... | ... |
| 2020-10-30 | 15185 |
| 2020-10-31 | 6243 |
| 2020-11-01 | 2660 |
| 2020-11-02 | 13345 |
| 2020-11-03 | 11167 |
248 rows × 1 columns
Check the types of the columns:
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 248 entries, 2020-03-01 to 2020-11-03
Data columns (total 1 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cases 248 non-null int64
dtypes: int64(1)
memory usage: 3.9 KB
Robert Koch Institute method
A basic method to calculate the effective reproduction number is described (among others) in this blogpost. I included the relevant paragraph:
In a recent report (an der Heiden and Hamouda 2020) the RKI described their method for computing R as part of the COVID-19 outbreak as follows (p. 13): For a constant generation time of 4 days, one obtains R as the ratio of new infections in two consecutive time periods each consisting of 4 days. Mathematically, this estimation could be formulated as part of a statistical model:
$$y_{s+4} | y_{s} \sim Po(R \cdot y_{s}), s= 1,2,3,4$$
where $y_{1}, \ldots, y_{4}$ are considered as fixed. From this we obtain
$$\hat{R}{RKI} = \sum$$}^{4} y_{s+4} / \sum_{s=1}^{4} y_{s
Somewhat arbitrary, we denote by $Re(t)$ the above estimate for R when $s=1$ corresponds to time $t-8$, i.e. we assign the obtained value to the last of the 8 values used in the computation.
In Python, we define a lambda function that we apply on a rolling window. Since indexes start from zero, we calculate:
$$\hat{R}{RKI} = \sum$$}^{3} y_{s+4} / \sum_{s=0}^{3} y_{s
| cases | |
|---|---|
| date | |
| 2020-03-01 | NaN |
| 2020-03-02 | NaN |
| 2020-03-03 | NaN |
| 2020-03-04 | NaN |
| 2020-03-05 | NaN |
| ... | ... |
| 2020-10-30 | 1.273703 |
| 2020-10-31 | 0.929291 |
| 2020-11-01 | 0.601838 |
| 2020-11-02 | 0.499806 |
| 2020-11-03 | 0.475685 |
248 rows × 1 columns
The first values are Nan because the window is in the past. If we plot the result, it looks like this:
ax = df.rolling(8).apply(rt).plot(figsize=(16,4), label='Re(t)')
ax.set(ylabel='Re(t)', title='Effective reproduction number estimated with RKI method')
ax.legend(['Re(t)']);

To avoid the spikes due to weekend reporting issue, I first applied a rolling mean on a window of 7 days:
ax = df.rolling(7).mean().rolling(8).apply(rt).plot(figsize=(16,4), label='Re(t)')
ax.set(ylabel='Re(t)', title='Effective reproduction number estimated with RKI method after rolling mean on window of 7 days')
ax.legend(['Re(t)']);

Interactive visualisation in Altair
import altair as alt
alt.Chart(df.rolling(7).mean().rolling(8).apply(rt).fillna(0).reset_index()).mark_line().encode(
x=alt.X('date:T'),
y=alt.Y('cases', title='Re(t)'),
tooltip=['date:T', alt.Tooltip('cases', format='.2f')]
).transform_filter(
alt.datum.date > alt.expr.toDate('2020-03-13')
).properties(
width=600,
title='Effective reproduction number in Belgium based on Robert-Koch Institute method'
)
Making the final visualisation in Altair
In the interactive Altair figure below, we show the $Re(t)$ for the last 14 days. We reduce the rolling mean window to three to see faster reactions.
#collapse
df_plot = df.rolling(7).mean().rolling(8).apply(rt).fillna(0).reset_index()
last_value = str(df_plot.iloc[-1]['cases'].round(2)) + ' ↓'
first_value = str(df_plot[df_plot['date'] == '2020-10-21'].iloc[0]['cases'].round(2)) # + ' ↑'
today_minus_15 = datetime.datetime.today() + relativedelta(days=-15)
today_minus_15_str = today_minus_15.strftime("%Y-%m-%d")
line = alt.Chart(df_plot).mark_line(point=True).encode(
x=alt.X('date:T', axis=alt.Axis(title='Datum', grid=False)),
y=alt.Y('cases', axis=alt.Axis(title='Re(t)', grid=False, labels=False, titlePadding=40)),
tooltip=['date:T', alt.Tooltip('cases', title='Re(t)', format='.2f')]
).transform_filter(
alt.datum.date > alt.expr.toDate(today_minus_15_str)
).properties(
width=600,
height=100
)
hline = alt.Chart(pd.DataFrame({'cases': [1]})).mark_rule().encode(y='cases')
label_right = alt.Chart(df_plot).mark_text(
align='left', dx=5, dy=-10 , size=15
).encode(
x=alt.X('max(date):T', title=None),
text=alt.value(last_value),
)
label_left = alt.Chart(df_plot).mark_text(
align='right', dx=-5, dy=-40, size=15
).encode(
x=alt.X('min(date):T', title=None),
text=alt.value(first_value),
).transform_filter(
alt.datum.date > alt.expr.toDate(today_minus_15_str)
)
source = alt.Chart(
{"values": [{"text": "Data source: Sciensano"}]}
).mark_text(size=12, align='left', dx=-57).encode(
text="text:N"
)
alt.vconcat(line + label_left + label_right + hline, source).configure(
background='#D9E9F0'
).configure_view(
stroke=None, # Remove box around graph
).configure_axisY(
ticks=False,
grid=False,
domain=False
).configure_axisX(
grid=False,
domain=False
).properties(title={
"text": ['Effective reproduction number for the last 14 days in Belgium'],
"subtitle": [f'Estimation based on the number of cases until {today_minus_two.strftime("%Y-%m-%d")} after example of Robert Koch Institute with serial interval of 4'],
}
)
# .configure_axisY(
# labelPadding=50,
# )
To check the calculation, here are the last for values for the number of cases after applying the mean window of 7:
| cases | |
|---|---|
| date | |
| 2020-10-27 | 16067.571429 |
| 2020-10-28 | 16135.857143 |
| 2020-10-29 | 15744.571429 |
| 2020-10-30 | 15218.000000 |
Those must be added together:
cases 63166.0
dtype: float64
And here are the four values, starting four days ago:
| cases | |
|---|---|
| date | |
| 2020-10-31 | 14459.428571 |
| 2020-11-01 | 14140.428571 |
| 2020-11-02 | 13213.428571 |
| 2020-11-03 | 11641.428571 |
These are added together:
cases 53454.714286
dtype: float64
And now we divide those two sums to get the $Re(t)$ of 2020-11-03:
cases 0.846258
dtype: float64
This matches (as expected) the value in the graph. Let's compare with three other sources:
- Alas it does not match the calculation reported by Bart Mesuere on 2020-11-03 based on the RKI model that reports 0.96:
twitter: https://twitter.com/BartMesuere/status/1323519613855059968
-
Also, the more elaborated model from rtliveglobal is not yet that optimistic. Mind that model rtlive start estimating the $Re(t)$ from the number of tests instead of the number of cases. It might be that other reporting delays are involved.
-
epiforecast.io is already below 1 since beginning of November.
Another possiblity is that I made somewhere a mistake. If you spot it, please let me know.
My talk at data science leuven
Links to video and slides of the talk and thanking people.
Talk material
On 23 April 2020, I was invited for a talk at Data science Leuven. I talked about how you can explore and explain the results of a clustering exercise. The target audience are data scientists that that have notions of how to cluster data and that want to improve their skills.
The video is recorded on Youtube:
youtube: https://youtu.be/hk0arqhcX9U?t=3570
You can see the slides here: slides
The talk itself is based on this notebook that I published on this blog yesterday and that I used to demo during the talk.
The host of the conference was Istvan Hajnal. He tweeted the following:
twitter: https://twitter.com/dsleuven/status/1253391470444371968
He also took the R out of my family name NachteRgaele. Troubles with R, it's becoming a story of my life... 

Standing on the shoulders of the giants
This talks was not possible without the awesome Altair visualisation library made by Jake VanderPlas. Secondly, it builds upon the open source Shap library made by Scott Lundberg. Those two libraries had a major impact on my daily work as datascientist at Colruyt group. They inspired me in trying to give back to the open source community with this talk.
If you want to learn how to use Altair I recommend the tutorial made by Vincent Warmerdam on his calm code site: https://calmcode.io/altair/introduction.htm
I would also like to thank my collegues at work who endured the dry-run of this talk and who made the suggestion to try to use a classifier to explain the clustering result. Top team!
Awesome fastpages
Finally, this blog is build with the awesome fastpages. I can now share a rendered Jupyter notebook, with working interactive demos, that can be opened in My binder or Google Colab with one click on a button. This means that readers can directly tinker around with the code and methods discussed in the talk. All you need is a browser and an internet connection. So thank you Jeremy Howard, Hamel Husain, and the fastdotai team for pulling this off. Thank you Hamel Husain for your Github Actions. I will cast for two how awesome this all is.
Regional covid-19 mortality in Belgium per gender and age
Combines the mortality number of the last 10 year with those of covid-19 this year.
df_inhab = pd.read_excel('https://statbel.fgov.be/sites/default/files/files/opendata/bevolking%20naar%20woonplaats%2C%20nationaliteit%20burgelijke%20staat%20%2C%20leeftijd%20en%20geslacht/TF_SOC_POP_STRUCT_2019.xlsx')
| CD_REFNIS | TX_DESCR_NL | TX_DESCR_FR | CD_DSTR_REFNIS | TX_ADM_DSTR_DESCR_NL | TX_ADM_DSTR_DESCR_FR | CD_PROV_REFNIS | TX_PROV_DESCR_NL | TX_PROV_DESCR_FR | CD_RGN_REFNIS | TX_RGN_DESCR_NL | TX_RGN_DESCR_FR | CD_SEX | CD_NATLTY | TX_NATLTY_NL | TX_NATLTY_FR | CD_CIV_STS | TX_CIV_STS_NL | TX_CIV_STS_FR | CD_AGE | MS_POPULATION | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 69 | 11 |
| 1 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 80 | 3 |
| 2 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | M | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 30 | 2 |
| 3 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 48 | 26 |
| 4 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 76 | 2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 463376 | 93090 | Viroinval | Viroinval | 93000 | Arrondissement Philippeville | Arrondissement de Philippeville | 90000.0 | Provincie Namen | Province de Namur | 3000 | Waals Gewest | Région wallonne | F | BEL | Belgen | Belges | 3 | Weduwstaat | Veuf | 73 | 10 |
| 463377 | 93090 | Viroinval | Viroinval | 93000 | Arrondissement Philippeville | Arrondissement de Philippeville | 90000.0 | Provincie Namen | Province de Namur | 3000 | Waals Gewest | Région wallonne | M | BEL | Belgen | Belges | 3 | Weduwstaat | Veuf | 64 | 1 |
| 463378 | 93090 | Viroinval | Viroinval | 93000 | Arrondissement Philippeville | Arrondissement de Philippeville | 90000.0 | Provincie Namen | Province de Namur | 3000 | Waals Gewest | Région wallonne | M | BEL | Belgen | Belges | 3 | Weduwstaat | Veuf | 86 | 3 |
| 463379 | 93090 | Viroinval | Viroinval | 93000 | Arrondissement Philippeville | Arrondissement de Philippeville | 90000.0 | Provincie Namen | Province de Namur | 3000 | Waals Gewest | Région wallonne | M | ETR | niet-Belgen | non-Belges | 3 | Weduwstaat | Veuf | 74 | 1 |
| 463380 | 93090 | Viroinval | Viroinval | 93000 | Arrondissement Philippeville | Arrondissement de Philippeville | 90000.0 | Provincie Namen | Province de Namur | 3000 | Waals Gewest | Région wallonne | M | BEL | Belgen | Belges | 3 | Weduwstaat | Veuf | 52 | 1 |
463381 rows × 21 columns
array(['Provincie Antwerpen', 'Provincie Vlaams-Brabant',
'Provincie Waals-Brabant', 'Provincie West-Vlaanderen',
'Provincie Oost-Vlaanderen', 'Provincie Henegouwen',
'Provincie Luik', 'Provincie Limburg', 'Provincie Luxemburg',
'Provincie Namen'], dtype=object)
array(['Brussels', 'Liège', 'Limburg', 'OostVlaanderen', 'VlaamsBrabant',
'Antwerpen', 'WestVlaanderen', 'BrabantWallon', 'Hainaut', 'Namur',
nan, 'Luxembourg'], dtype=object)
['Antwerpen',
'Vlaams-Brabant',
'Waals-Brabant',
'West-Vlaanderen',
'Oost-Vlaanderen',
'Henegouwen',
'Luik',
'Limburg',
'Luxemburg',
'Namen']
map_statbel_provence_to_sc_provence = {'Provincie Antwerpen':'Antwerpen', 'Provincie Vlaams-Brabant':'VlaamsBrabant',
'Provincie Waals-Brabant':'BrabantWallon', 'Provincie West-Vlaanderen':'WestVlaanderen',
'Provincie Oost-Vlaanderen':'OostVlaanderen', 'Provincie Henegouwen':'Hainaut',
'Provincie Luik':'Liège', 'Provincie Limburg':'Limburg', 'Provincie Luxemburg':'Luxembourg',
'Provincie Namen':'Namur'}
array(['10-19', '20-29', '30-39', '40-49', '50-59', '70-79', '60-69',
'0-9', '90+', '80-89', nan], dtype=object)
df_inhab['AGEGROUP'] =pd.cut(df_inhab['CD_AGE'], bins=[0,10,20,30,40,50,60,70,80,90,200], labels=['0-9','10-19','20-29','30-39','40-49','50-59','60-69','70-79','80-89','90+'], include_lowest=True)
df_inhab_gender_prov = df_inhab.groupby(['sc_provence', 'CD_SEX', 'AGEGROUP'])['MS_POPULATION'].sum().reset_index()
df_inhab_gender_prov_cases = pd.merge(df_inhab_gender_prov, df_tot_sc.dropna(), left_on=['sc_provence', 'AGEGROUP', 'CD_SEX'], right_on=['PROVINCE', 'AGEGROUP', 'SEX'])
| sc_provence | CD_SEX | AGEGROUP | MS_POPULATION | DATE | PROVINCE | REGION | SEX | CASES | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Antwerpen | F | 0-9 | 113851 | 2020-03-05 | Antwerpen | Flanders | F | 1 |
| 1 | Antwerpen | F | 0-9 | 113851 | 2020-03-18 | Antwerpen | Flanders | F | 1 |
| 2 | Antwerpen | F | 0-9 | 113851 | 2020-03-26 | Antwerpen | Flanders | F | 1 |
| 3 | Antwerpen | F | 0-9 | 113851 | 2020-03-30 | Antwerpen | Flanders | F | 1 |
| 4 | Antwerpen | F | 0-9 | 113851 | 2020-04-03 | Antwerpen | Flanders | F | 1 |
df_plot = df_inhab_gender_prov_cases.groupby(['SEX', 'AGEGROUP', 'PROVINCE']).agg(CASES = ('CASES', 'sum'), MS_POPULATION=('MS_POPULATION', 'first')).reset_index()
df_plot
| SEX | AGEGROUP | PROVINCE | CASES | MS_POPULATION | |
|---|---|---|---|---|---|
| 0 | F | 0-9 | Antwerpen | 9 | 113851 |
| 1 | F | 0-9 | BrabantWallon | 3 | 23744 |
| 2 | F | 0-9 | Hainaut | 11 | 81075 |
| 3 | F | 0-9 | Limburg | 11 | 48102 |
| 4 | F | 0-9 | Liège | 19 | 67479 |
| ... | ... | ... | ... | ... | ... |
| 195 | M | 90+ | Luxembourg | 17 | 469 |
| 196 | M | 90+ | Namur | 27 | 827 |
| 197 | M | 90+ | OostVlaanderen | 102 | 3105 |
| 198 | M | 90+ | VlaamsBrabant | 129 | 2611 |
| 199 | M | 90+ | WestVlaanderen | 121 | 3292 |
200 rows × 5 columns
array(['Antwerpen', 'BrabantWallon', 'Hainaut', 'Limburg', 'Liège',
'Luxembourg', 'Namur', 'OostVlaanderen', 'VlaamsBrabant',
'WestVlaanderen'], dtype=object)
alt.Chart(df_plot).mark_bar().encode(x='AGEGROUP:N', y='percentage', color='SEX:N', column='PROVINCE:N')
Let's add a colorscale the makes the male blue and female number pink.
alt.Chart(df_plot).mark_bar().encode(
x='AGEGROUP:N',
y='percentage',
color=alt.Color('SEX:N', scale=color_scale, legend=None),
column='PROVINCE:N')
The graph's get to wide. Let's use faceting to make two rows.
Inspired and based on https://altair-viz.github.io/gallery/us_population_pyramid_over_time.html
#slider = alt.binding_range(min=1850, max=2000, step=10)
# select_province = alt.selection_single(name='PROVINCE', fields=['PROVINCE'],
# bind=slider, init={'PROVINCE': 'Antwerpen'})
color_scale = alt.Scale(domain=['Male', 'Female'],
range=['#1f77b4', '#e377c2'])
select_province = alt.selection_multi(fields=['PROVINCE'], bind='legend')
base = alt.Chart(df_plot).add_selection(
select_province
).transform_filter(
select_province
).transform_calculate(
gender=alt.expr.if_(alt.datum.SEX == 'M', 'Male', 'Female')
).properties(
width=250
)
left = base.transform_filter(
alt.datum.gender == 'Female'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
x=alt.X('percentage:Q', axis=alt.Axis(format='.0%'),
title='Percentage',
sort=alt.SortOrder('descending'),
),
color=alt.Color('gender:N', scale=color_scale, legend=None),
).mark_bar().properties(title='Female')
middle = base.encode(
y=alt.Y('AGEGROUP:O', axis=None),
text=alt.Text('AGEGROUP:O'),
).mark_text().properties(width=20)
right = base.transform_filter(
alt.datum.gender == 'Male'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
x=alt.X('percentage:Q', title='Percentage', axis=alt.Axis(format='.0%'),),
color=alt.Color('gender:N', scale=color_scale, legend=None)
).mark_bar().properties(title='Male')
# legend = alt.Chart(df_plot).mark_text().encode(
# y=alt.Y('PROVINCE:N', axis=None),
# text=alt.Text('PROVINCE:N'),
# color=alt.Color('PROVINCE:N', legend=alt.Legend(title="Provincie"))
# )
alt.concat(left, middle, right, spacing=5)
#legend=alt.Legend(title="Species by color")
provinces = df_plot['PROVINCE'].unique()
select_province = alt.selection_single(
name='Select', # name the selection 'Select'
fields=['PROVINCE'], # limit selection to the Major_Genre field
init={'PROVINCE': 'Antwerpen'}, # use first genre entry as initial value
bind=alt.binding_select(options=provinces) # bind to a menu of unique provence values
)
base = alt.Chart(df_plot).add_selection(
select_province
).transform_filter(
select_province
).transform_calculate(
gender=alt.expr.if_(alt.datum.SEX == 'M', 'Male', 'Female')
).properties(
width=250
)
left = base.transform_filter(
alt.datum.gender == 'Female'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
x=alt.X('percentage:Q', axis=alt.Axis(format='.0%'),
title='Percentage',
sort=alt.SortOrder('descending'),
scale=alt.Scale(domain=(0.0, 0.1), clamp=True)
),
color=alt.Color('gender:N', scale=color_scale, legend=None),
tooltip=[alt.Tooltip('percentage', format='.1%')]
).mark_bar().properties(title='Female')
middle = base.encode(
y=alt.Y('AGEGROUP:O', axis=None),
text=alt.Text('AGEGROUP:O'),
).mark_text().properties(width=20)
right = base.transform_filter(
alt.datum.gender == 'Male'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
x=alt.X('percentage:Q', title='Percentage', axis=alt.Axis(format='.1%'), scale=alt.Scale(domain=(0.0, 0.1), clamp=True)),
color=alt.Color('gender:N', scale=color_scale, legend=None),
tooltip=[alt.Tooltip('percentage', format='.1%')]
).mark_bar().properties(title='Male')
alt.concat(left, middle, right, spacing=5).properties(title='Percentage of covid-19 cases per province, gender and age grup in Belgium')
Mortality
# https://epistat.wiv-isp.be/covid/
# Dataset of mortality by date, age, sex, and region
df_dead_sc = pd.read_csv('https://epistat.sciensano.be/Data/COVID19BE_MORT.csv')
| DATE | REGION | AGEGROUP | SEX | DEATHS | |
|---|---|---|---|---|---|
| 0 | 2020-03-10 | Brussels | 85+ | F | 1 |
| 1 | 2020-03-11 | Flanders | 85+ | F | 1 |
| 2 | 2020-03-11 | Brussels | 75-84 | M | 1 |
| 3 | 2020-03-11 | Brussels | 85+ | F | 1 |
| 4 | 2020-03-12 | Brussels | 75-84 | M | 1 |
Wallonia 291
Flanders 275
Brussels 271
Name: REGION, dtype: int64
85+ 223
75-84 205
65-74 179
45-64 132
25-44 19
0-24 1
Name: AGEGROUP, dtype: int64
df_inhab['AGEGROUP_sc'] =pd.cut(df_inhab['CD_AGE'], bins=[0,24,44,64,74,84,200], labels=['0-24','25-44','45-64','65-74','75-84','85+'], include_lowest=True)
| lowest_age | highest_age | |
|---|---|---|
| AGEGROUP_sc | ||
| 0-24 | 0 | 24 |
| 25-44 | 25 | 44 |
| 45-64 | 45 | 64 |
| 65-74 | 65 | 74 |
| 75-84 | 75 | 84 |
| 85+ | 85 | 110 |
| CD_REFNIS | TX_DESCR_NL | TX_DESCR_FR | CD_DSTR_REFNIS | TX_ADM_DSTR_DESCR_NL | TX_ADM_DSTR_DESCR_FR | CD_PROV_REFNIS | TX_PROV_DESCR_NL | TX_PROV_DESCR_FR | CD_RGN_REFNIS | TX_RGN_DESCR_NL | TX_RGN_DESCR_FR | CD_SEX | CD_NATLTY | TX_NATLTY_NL | TX_NATLTY_FR | CD_CIV_STS | TX_CIV_STS_NL | TX_CIV_STS_FR | CD_AGE | MS_POPULATION | sc_provence | AGEGROUP | AGEGROUP_sc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 69 | 11 | Antwerpen | 60-69 | 65-74 |
| 1 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 80 | 3 | Antwerpen | 70-79 | 75-84 |
| 2 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | M | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 30 | 2 | Antwerpen | 20-29 | 25-44 |
| 3 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 48 | 26 | Antwerpen | 40-49 | 45-64 |
| 4 | 11001 | Aartselaar | Aartselaar | 11000 | Arrondissement Antwerpen | Arrondissement d’Anvers | 10000.0 | Provincie Antwerpen | Province d’Anvers | 2000 | Vlaams Gewest | Région flamande | F | BEL | Belgen | Belges | 4 | Gescheiden | Divorcé | 76 | 2 | Antwerpen | 70-79 | 75-84 |
array(['Brussels', 'Flanders', 'Wallonia'], dtype=object)
Vlaams Gewest 242865
Waals Gewest 199003
Brussels Hoofdstedelijk Gewest 21513
Name: TX_RGN_DESCR_NL, dtype: int64
df_inhab_gender_prov = df_inhab.groupby(['TX_RGN_DESCR_NL', 'CD_SEX', 'AGEGROUP_sc'])['MS_POPULATION'].sum().reset_index()
region_sc_to_region_inhad = {'Flanders':'Vlaams Gewest', 'Wallonia':'Waals Gewest', 'Brussels':'Brussels Hoofdstedelijk Gewest'}
TX_RGN_DESCR_NL AGEGROUP SEX
Brussels Hoofdstedelijk Gewest 25-44 F 1
M 4
45-64 F 21
M 43
65-74 F 42
M 71
75-84 F 128
M 170
85+ F 270
M 186
Vlaams Gewest 0-24 F 1
25-44 F 2
M 3
45-64 F 27
M 63
65-74 F 67
M 130
75-84 F 199
M 335
85+ F 232
M 309
Waals Gewest 25-44 F 5
M 4
45-64 F 41
M 89
65-74 F 98
M 186
75-84 F 290
M 300
85+ F 704
M 421
Name: DEATHS, dtype: int64
df_dead_sc_region_agegroup_gender = df_dead_sc.groupby(['TX_RGN_DESCR_NL', 'AGEGROUP', 'SEX'])['DEATHS'].sum().reset_index()
df_inhab_gender_prov_deaths = pd.merge(df_inhab_gender_prov, df_dead_sc_region_agegroup_gender, left_on=['TX_RGN_DESCR_NL', 'AGEGROUP_sc', 'CD_SEX'], right_on=['TX_RGN_DESCR_NL', 'AGEGROUP', 'SEX'])
9077403
4442
| TX_RGN_DESCR_NL | CD_SEX | AGEGROUP_sc | MS_POPULATION | AGEGROUP | SEX | DEATHS | |
|---|---|---|---|---|---|---|---|
| 0 | Brussels Hoofdstedelijk Gewest | F | 25-44 | 197579 | 25-44 | F | 1 |
| 1 | Brussels Hoofdstedelijk Gewest | F | 45-64 | 137628 | 45-64 | F | 21 |
| 2 | Brussels Hoofdstedelijk Gewest | F | 65-74 | 45214 | 65-74 | F | 42 |
| 3 | Brussels Hoofdstedelijk Gewest | F | 75-84 | 30059 | 75-84 | F | 128 |
| 4 | Brussels Hoofdstedelijk Gewest | F | 85+ | 18811 | 85+ | F | 270 |
| 5 | Brussels Hoofdstedelijk Gewest | M | 25-44 | 194988 | 25-44 | M | 4 |
| 6 | Brussels Hoofdstedelijk Gewest | M | 45-64 | 140348 | 45-64 | M | 43 |
| 7 | Brussels Hoofdstedelijk Gewest | M | 65-74 | 36698 | 65-74 | M | 71 |
| 8 | Brussels Hoofdstedelijk Gewest | M | 75-84 | 19969 | 75-84 | M | 170 |
| 9 | Brussels Hoofdstedelijk Gewest | M | 85+ | 7918 | 85+ | M | 186 |
| 10 | Vlaams Gewest | F | 0-24 | 874891 | 0-24 | F | 1 |
| 11 | Vlaams Gewest | F | 25-44 | 820036 | 25-44 | F | 2 |
| 12 | Vlaams Gewest | F | 45-64 | 901554 | 45-64 | F | 27 |
| 13 | Vlaams Gewest | F | 65-74 | 353925 | 65-74 | F | 67 |
| 14 | Vlaams Gewest | F | 75-84 | 245981 | 75-84 | F | 199 |
| 15 | Vlaams Gewest | F | 85+ | 132649 | 85+ | F | 232 |
| 16 | Vlaams Gewest | M | 25-44 | 827281 | 25-44 | M | 3 |
| 17 | Vlaams Gewest | M | 45-64 | 917008 | 45-64 | M | 63 |
| 18 | Vlaams Gewest | M | 65-74 | 336242 | 65-74 | M | 130 |
| 19 | Vlaams Gewest | M | 75-84 | 193576 | 75-84 | M | 335 |
| 20 | Vlaams Gewest | M | 85+ | 69678 | 85+ | M | 309 |
| 21 | Waals Gewest | F | 25-44 | 457356 | 25-44 | F | 5 |
| 22 | Waals Gewest | F | 45-64 | 496668 | 45-64 | F | 41 |
| 23 | Waals Gewest | F | 65-74 | 199422 | 65-74 | F | 98 |
| 24 | Waals Gewest | F | 75-84 | 118224 | 75-84 | F | 290 |
| 25 | Waals Gewest | F | 85+ | 68502 | 85+ | F | 704 |
| 26 | Waals Gewest | M | 25-44 | 459444 | 25-44 | M | 4 |
| 27 | Waals Gewest | M | 45-64 | 487322 | 45-64 | M | 89 |
| 28 | Waals Gewest | M | 65-74 | 175508 | 65-74 | M | 186 |
| 29 | Waals Gewest | M | 75-84 | 82876 | 75-84 | M | 300 |
| 30 | Waals Gewest | M | 85+ | 30048 | 85+ | M | 421 |
df_inhab_gender_prov_deaths['percentage'] = df_inhab_gender_prov_deaths['DEATHS']/df_inhab_gender_prov_deaths['MS_POPULATION']
regions = df_plot['TX_RGN_DESCR_NL'].unique()
select_province = alt.selection_single(
name='Select', # name the selection 'Select'
fields=['TX_RGN_DESCR_NL'], # limit selection to the Major_Genre field
init={'TX_RGN_DESCR_NL': 'Vlaams Gewest'}, # use first genre entry as initial value
bind=alt.binding_select(options=regions) # bind to a menu of unique provence values
)
base = alt.Chart(df_plot).add_selection(
select_province
).transform_filter(
select_province
).transform_calculate(
gender=alt.expr.if_(alt.datum.SEX == 'M', 'Male', 'Female')
).properties(
width=250
)
left = base.transform_filter(
alt.datum.gender == 'Female'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
x=alt.X('percentage:Q', axis=alt.Axis(format='.2%'),
title='Percentage',
sort=alt.SortOrder('descending'),
# scale=alt.Scale(domain=(0.0, 0.02), clamp=True)
),
color=alt.Color('gender:N', scale=color_scale, legend=None),
tooltip=[alt.Tooltip('percentage', format='.2%')]
).mark_bar().properties(title='Female')
middle = base.encode(
y=alt.Y('AGEGROUP:O', axis=None),
text=alt.Text('AGEGROUP:O'),
).mark_text().properties(width=20)
right = base.transform_filter(
alt.datum.gender == 'Male'
).encode(
y=alt.Y('AGEGROUP:O', axis=None),
# x=alt.X('percentage:Q', title='Percentage', axis=alt.Axis(format='.2%'), scale=alt.Scale(domain=(0.0, 0.02), clamp=True)),
x=alt.X('percentage:Q', title='Percentage', axis=alt.Axis(format='.2%')),
color=alt.Color('gender:N', scale=color_scale, legend=None),
tooltip=[alt.Tooltip('percentage', format='.2%')]
).mark_bar().properties(title='Male')
alt.concat(left, middle, right, spacing=5).properties(title='Percentage of covid-19 deaths per province, gender and age group relative to number of inhabitants in Belgium')
Daily covid-19 Deaths compared to average deaths the last 10 years
"In this blogpost we try to get an idea of how many extra deaths we have in Belgium due to covid-19 compared to the average we had the last 10 years."
The number of deadths per day from 2008 until 2018 can obtained from Statbel, the Belgium federal bureau of statistics:
df = pd.read_excel('https://statbel.fgov.be/sites/default/files/files/opendata/bevolking/TF_DEATHS.xlsx') # , skiprows=5, sheet_name=sheetnames
| DT_DATE | MS_NUM_DEATHS | |
|---|---|---|
| 0 | 2008-01-01 | 342 |
| 1 | 2008-01-02 | 348 |
| 2 | 2008-01-03 | 340 |
| 3 | 2008-01-04 | 349 |
| 4 | 2008-01-05 | 348 |
# Let's make a quick plot
alt.Chart(df_plot).mark_line().encode(x='Dag', y='MS_NUM_DEATHS').properties(width=600)
The John Hopkings University CSSE keeps track of the number of covid-19 deadths per day and country in a github repository: https://github.com/CSSEGISandData/COVID-19. We can easily obtain this data by reading it from github and filter out the cases for Belgium.
deaths_url = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv'
deaths = pd.read_csv(deaths_url, sep=',')
Filter out Belgium
Inspect how the data is stored
| Province/State | Country/Region | Lat | Long | 1/22/20 | 1/23/20 | 1/24/20 | 1/25/20 | 1/26/20 | 1/27/20 | ... | 4/9/20 | 4/10/20 | 4/11/20 | 4/12/20 | 4/13/20 | 4/14/20 | 4/15/20 | 4/16/20 | 4/17/20 | 4/18/20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 23 | NaN | Belgium | 50.8333 | 4.0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 2523 | 3019 | 3346 | 3600 | 3903 | 4157 | 4440 | 4857 | 5163 | 5453 |
1 rows × 92 columns
Create dateframe for plotting
df_deaths = pd.DataFrame(data={'Datum':pd.to_datetime(deaths_be.columns[4:]), 'Overlijdens':deaths_be.iloc[0].values[4:]})
Check for Nan's
0
We need to do some type convertions. We cast 'Overlijdens' to integer. Next, we add the number of the day.
df_deaths['Overlijdens'] = df_deaths['Overlijdens'].astype(int)
df_deaths['Dag'] = df_deaths['Datum'].dt.dayofyear
Plot the data:
Calculate the day-by-day change
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 88 entries, 0 to 87
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Datum 88 non-null datetime64[ns]
1 Overlijdens 88 non-null int32
2 Dag 88 non-null int64
3 Nieuwe covid-19 Sterfgevallen 87 non-null float64
dtypes: datetime64[ns](1), float64(1), int32(1), int64(1)
memory usage: 2.5 KB
Plot covid-19 deaths in Belgium according to JHU CSSE. The plot shows a tooltip if you hover over the points.
dead_covid= alt.Chart(df_deaths).mark_line(point=True).encode(
x=alt.X('Dag',scale=alt.Scale(domain=(1, 110), clamp=True)),
y='Nieuwe covid-19 Sterfgevallen',
color=alt.ColorValue('red'),
tooltip=['Dag', 'Nieuwe covid-19 Sterfgevallen'])
dead_covid
Now we add average deaths per day in the last 10 year to the plot.
Take quick look to the datatable:
| DT_DATE | MS_NUM_DEATHS | Jaar | Dag | |
|---|---|---|---|---|
| 0 | 2008-01-01 | 342 | 2008 | 1 |
| 1 | 2008-01-02 | 348 | 2008 | 2 |
| 2 | 2008-01-03 | 340 | 2008 | 3 |
| 3 | 2008-01-04 | 349 | 2008 | 4 |
| 4 | 2008-01-05 | 348 | 2008 | 5 |
The column 'DT_DATE' is a string. We convert it to a datatime so we can add it to the tooltip.
Now we are prepared to make the final graph. We use the Altair mark_errorband(extend='ci') to bootstrap 95% confidence band around the average number of deaths per day.
line = alt.Chart(df).mark_line().encode(
x=alt.X('Dag', scale=alt.Scale(
domain=(1, 120),
clamp=True
)),
y='mean(MS_NUM_DEATHS)'
)
# Bootstrapped 95% confidence interval
band = alt.Chart(df).mark_errorband(extent='ci').encode(
x=alt.X('Dag', scale=alt.Scale(domain=(1, 120), clamp=True)),
y=alt.Y('MS_NUM_DEATHS', title='Overlijdens per dag'),
)
dead_covid= alt.Chart(df_deaths).mark_line(point=True).encode(
x=alt.X('Dag',scale=alt.Scale(domain=(1, 120), clamp=True)),
y='Nieuwe covid-19 Sterfgevallen',
color=alt.ColorValue('red'),
tooltip=['Dag', 'Nieuwe covid-19 Sterfgevallen', 'Datum']
)
(band + line + dead_covid).properties(width=1024, title='Gemiddeld aantal overlijdens over 10 jaar versus overlijdens door covid-19 in Belgie')
Source date from sciensano
In this section, we compare the graph obtained with data obtained from sciensano.
| DATE | REGION | AGEGROUP | SEX | DEATHS | |
|---|---|---|---|---|---|
| 0 | 2020-03-10 | Brussels | 85+ | F | 1 |
| 1 | 2020-03-11 | Flanders | 85+ | F | 1 |
| 2 | 2020-03-11 | Brussels | 75-84 | M | 1 |
| 3 | 2020-03-11 | Brussels | 85+ | F | 1 |
| 4 | 2020-03-12 | Brussels | 75-84 | M | 1 |
df_dead_day = df_sc.groupby('DATE')['DEATHS'].sum().reset_index()
df_dead_day['Datum'] = pd.to_datetime(df_dead_day['DATE'])
df_dead_day['Dag'] = df_dead_day['Datum'].dt.dayofyear
line = alt.Chart(df).mark_line().encode(
x=alt.X('Dag', title='Dag van het jaar', scale=alt.Scale(
domain=(1, 120),
clamp=True
)),
y='mean(MS_NUM_DEATHS)'
)
# Bootstrapped 95% confidence interval
band = alt.Chart(df).mark_errorband(extent='ci').encode(
x=alt.X('Dag', scale=alt.Scale(domain=(1, 120), clamp=True)),
y=alt.Y('MS_NUM_DEATHS', title='Overlijdens per dag'),
)
dead_covid= alt.Chart(df_dead_day).mark_line(point=True).encode(
x=alt.X('Dag',scale=alt.Scale(domain=(1, 120), clamp=True)),
y='DEATHS',
color=alt.ColorValue('red'),
tooltip=['Dag', 'DEATHS', 'Datum']
)
(band + line + dead_covid).properties(width=750, title='Gemiddeld aantal overlijdens over 10 jaar versus overlijdens door covid-19 in Belgie')
Obviously, data form 16-17-18 April 2020 is not final yet. Also, the amounts are smaller then those from JHU.
Obtain more detail (for another blogpost...)
| DATE | PROVINCE | REGION | AGEGROUP | SEX | CASES | |
|---|---|---|---|---|---|---|
| 0 | 2020-03-01 | Brussels | Brussels | 10-19 | M | 1 |
| 1 | 2020-03-01 | Brussels | Brussels | 10-19 | F | 1 |
| 2 | 2020-03-01 | Brussels | Brussels | 20-29 | M | 1 |
| 3 | 2020-03-01 | Brussels | Brussels | 30-39 | F | 1 |
| 4 | 2020-03-01 | Brussels | Brussels | 40-49 | F | 1 |
| ... | ... | ... | ... | ... | ... | ... |
| 6875 | NaN | OostVlaanderen | Flanders | NaN | F | 4 |
| 6876 | NaN | VlaamsBrabant | Flanders | 40-49 | M | 3 |
| 6877 | NaN | VlaamsBrabant | Flanders | 40-49 | F | 2 |
| 6878 | NaN | VlaamsBrabant | Flanders | 50-59 | M | 1 |
| 6879 | NaN | WestVlaanderen | Flanders | 50-59 | M | 3 |
6880 rows × 6 columns
We know that there are a lot of reional differences:
| DATE | PROVINCE | CASES | |
|---|---|---|---|
| 0 | 2020-03-01 | Brussels | 6 |
| 1 | 2020-03-01 | Limburg | 1 |
| 2 | 2020-03-01 | Liège | 2 |
| 3 | 2020-03-01 | OostVlaanderen | 1 |
| 4 | 2020-03-01 | VlaamsBrabant | 6 |
| ... | ... | ... | ... |
| 505 | 2020-04-17 | OostVlaanderen | 44 |
| 506 | 2020-04-17 | VlaamsBrabant | 42 |
| 507 | 2020-04-17 | WestVlaanderen | 30 |
| 508 | 2020-04-18 | Brussels | 1 |
| 509 | 2020-04-18 | Hainaut | 1 |
510 rows × 3 columns
base = alt.Chart(df_plot, title='Number of cases in Belgium per day and province').mark_line(point=True).encode(
x=alt.X('DATE:T', title='Datum'),
y=alt.Y('CASES', title='Cases per day'),
color='PROVINCE',
tooltip=['DATE', 'CASES', 'PROVINCE']
).properties(width=600)
base
From the above graph we see a much lower number of cases in Luxembourg, Namur, Waals Brabant.
'pwd' is not recognized as an internal or external command,
operable program or batch file.
Volume in drive C is Windows
Volume Serial Number is 7808-E933
Directory of C:\Users\lnh6dt5\AppData\Local\Temp\Mxt121\tmp\home_lnh6dt5\blog\_notebooks
19/04/2020 14:14 <DIR> .
19/04/2020 14:14 <DIR> ..
19/04/2020 10:37 <DIR> .ipynb_checkpoints
19/04/2020 10:17 23.473 2020-01-28-Altair.ipynb
19/04/2020 10:34 9.228 2020-01-29-bullet-chart-altair.ipynb
19/04/2020 10:26 41.041 2020-02-15-breakins.ipynb
19/04/2020 09:43 30.573 2020-02-20-test.ipynb
19/04/2020 09:49 1.047 2020-04-18-first-test.ipynb
19/04/2020 14:14 1.237.674 2020-04-19-deads-last-ten-year-vs-covid.ipynb
19/04/2020 09:43 <DIR> my_icons
19/04/2020 09:43 771 README.md
7 File(s) 1.343.807 bytes
4 Dir(s) 89.905.336.320 bytes free
First test post
Testing a simple notebook for publishing with fastpages
Evolution of burglary in Leuven. Is the trend downwards ?
Evolution of burglary in Leuven. Is the trend downwards ?
The local police shared a graph with the number of break-ins in Leuven per year. The article shows a graph with a downwards trendline. Can we conclude that the number of breakins is showing a downward trend based on those numbers? Let's construct a dataframe with the data from the graph.
import numpy as np
import pandas as pd
import altair as alt
df = pd.DataFrame({'year_int':[y for y in range(2006, 2020)],
'breakins':[1133,834,953,891,1006,1218,992,1079,1266,1112,713,669,730,644]})
df['year'] = pd.to_datetime(df['year_int'], format='%Y')
points = alt.Chart(df).mark_line(point=True).encode(
x='year', y='breakins', tooltip='breakins'
)
points + points.transform_regression('year', 'breakins').mark_line(
color='green'
).properties(
title='Regression trend on the number breakins per year in Leuven'
)
The article claims that the number of breakins stabilizes the last years. Let's perform a local regression to check that.
# https://opendatascience.com/local-regression-in-python
# Loess: https://gist.github.com/AllenDowney/818f6153ef316aee80467c51faee80f8
points + points.transform_loess('year', 'breakins').mark_line(
color='green'
).properties(
title='Local regression trend on the number breakins per year in Leuven'
)
But what about the trend line? Are we sure the trend is negative ? Bring in the code based on the blogpost The hacker's guide to uncertainty estimates to estimate the uncertainty.:
# Code from: https://erikbern.com/2018/10/08/the-hackers-guide-to-uncertainty-estimates.html
import scipy.optimize
import random
def model(xs, k, m):
return k * xs + m
def neg_log_likelihood(tup, xs, ys):
# Since sigma > 0, we use use log(sigma) as the parameter instead.
# That way we have an unconstrained problem.
k, m, log_sigma = tup
sigma = np.exp(log_sigma)
delta = model(xs, k, m) - ys
return len(xs)/2*np.log(2*np.pi*sigma**2) + \
np.dot(delta, delta) / (2*sigma**2)
def confidence_bands(xs, ys, nr_bootstrap):
curves = []
xys = list(zip(xs, ys))
for i in range(nr_bootstrap):
# sample with replacement
bootstrap = [random.choice(xys) for _ in xys]
xs_bootstrap = np.array([x for x, y in bootstrap])
ys_bootstrap = np.array([y for x, y in bootstrap])
k_hat, m_hat, log_sigma_hat = scipy.optimize.minimize(
neg_log_likelihood, (0, 0, 0), args=(xs_bootstrap, ys_bootstrap)
).x
curves.append(
model(xs, k_hat, m_hat) +
# Note what's going on here: we're _adding_ the random term
# to the predictions!
np.exp(log_sigma_hat) * np.random.normal(size=xs.shape)
)
lo, hi = np.percentile(curves, (2.5, 97.5), axis=0)
return lo, hi
# Make a plot with a confidence band
df['lo'], df['hi'] = confidence_bands(df.index, df['breakins'], 100)
ci = alt.Chart(df).mark_area().encode(
x=alt.X('year:T', title=''),
y=alt.Y('lo:Q'),
y2=alt.Y2('hi:Q', title=''),
color=alt.value('lightblue'),
opacity=alt.value(0.6)
)
chart = alt.Chart(df).mark_line(point=True).encode(
x='year', y='breakins', tooltip='breakins'
)
ci + chart + chart.transform_regression('year', 'breakins').mark_line(
color='red'
).properties(
title='95% Confidence band of the number of breakins per year in Leuven'
)
On the above chart, we see that a possitive trend might be possible as well.
Linear regression
Let's perform a linear regression with statsmodel to calculate the confidence interval on the slope of the regression line.
Intercept 1096.314286
index -23.169231
dtype: float64
The most likely slope of the trend line is 23.17 breakins per year. But how sure are we that the trend is heading down ?
C:\Users\lnh6dt5\AppData\Local\Continuum\anaconda3\lib\site-packages\scipy\stats\stats.py:1535: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=14
"anyway, n=%i" % int(n))
| Dep. Variable: | breakins | R-squared: | 0.223 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.159 |
| Method: | Least Squares | F-statistic: | 3.451 |
| Date: | Sun, 19 Apr 2020 | Prob (F-statistic): | 0.0879 |
| Time: | 10:26:45 | Log-Likelihood: | -92.105 |
| No. Observations: | 14 | AIC: | 188.2 |
| Df Residuals: | 12 | BIC: | 189.5 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 1096.3143 | 95.396 | 11.492 | 0.000 | 888.465 | 1304.164 |
| index | -23.1692 | 12.472 | -1.858 | 0.088 | -50.344 | 4.006 |
| Omnibus: | 1.503 | Durbin-Watson: | 1.035 |
|---|---|---|---|
| Prob(Omnibus): | 0.472 | Jarque-Bera (JB): | 1.196 |
| Skew: | 0.577 | Prob(JB): | 0.550 |
| Kurtosis: | 2.153 | Cond. No. | 14.7 |
Warnings:[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
The analysis reveals that the slope of the best fitting regression line is 23 breakins less per year. However, the confidence interval of the trend is between -50.344 and 4.006. Also the p)value of the regression coefficient is 0.088. Meaning we have eight percent chance that the negative trend is by accident. Hence, based on the current data we are not 95% percent sure the trend is downwards. Hence we can not conclude, based on this data, that there is a negative trend. This corresponds with the width of the 95% certainty band drawn that allows for an upward trend line:
| 0 | 1 | |
|---|---|---|
| Intercept | 888.464586 | 1304.163986 |
| index | -50.344351 | 4.005889 |
y_low = results.params['Intercept'] # ?ost likely value of the intercept
y_high = results.params['Intercept'] + results.conf_int()[1]['index'] * df.shape[0] # Value of upward trend for the last year
df_upward_trend = pd.DataFrame({'year':[df['year'].min(), df['year'].max()],
'breakins':[y_low, y_high]})
possible_upwards_trend = alt.Chart(df_upward_trend).mark_line(
color='green',
strokeDash=[10,10]
).encode(
x='year:T',
y=alt.Y('breakins:Q',
title='Number of breakins per year')
)
points = alt.Chart(df).mark_line(point=True).encode(x='year', y='breakins', tooltip='breakins')
(ci + points + points.transform_regression('year', 'breakins').mark_line(color='red')
+ possible_upwards_trend).properties(
title='Trend analysis on the number of breakins per year in Leuven, Belgium'
)
In the above graph, we see that a slight positive trend (green dashed line) is in the 95% confidence band on the regression coefficient. We are not sure that the trend on the number of breakins is downwards.
Bullet chart in python Altair
How to make bullet charts with Altair
In the article "Bullet Charts - What Is It And How To Use It" I learned about Bullet charts. It's a specific kind of barchart that must convey the state of a measure or KPI. The goal is to see in a glance if the target is met. Here is an example bullet chart from the article:
# This causes issues to:
# from IPython.display import Image
# Image('https://jscharting.com/blog/bullet-charts/images/bullet_components.png')
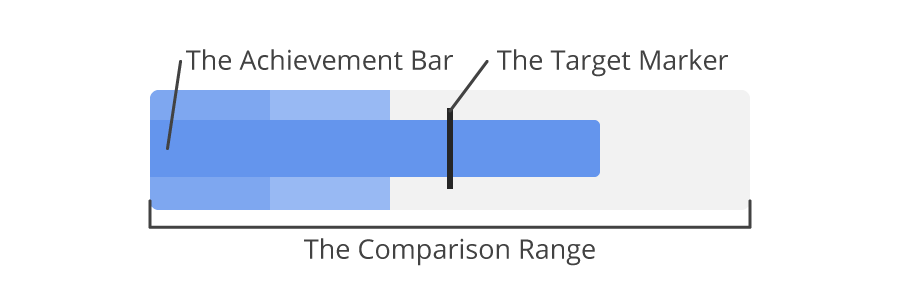
# <img src="https://jscharting.com/blog/bullet-charts/images/bullet_components.png" alt="Bullet chart" style="width: 200px;"/>
Below is some Python code that generates bullets graphs using the Altair library.
import altair as alt
import pandas as pd
df = pd.DataFrame.from_records([
{"title":"Revenue","subtitle":"US$, in thousands","ranges":[150,225,300],"measures":[220,270],"markers":[250]},
{"title":"Profit","subtitle":"%","ranges":[20,25,30],"measures":[21,23],"markers":[26]},
{"title":"Order Size","subtitle":"US$, average","ranges":[350,500,600],"measures":[100,320],"markers":[550]},
{"title":"New Customers","subtitle":"count","ranges":[1400,2000,2500],"measures":[1000,1650],"markers":[2100]},
{"title":"Satisfaction","subtitle":"out of 5","ranges":[3.5,4.25,5],"measures":[3.2,4.7],"markers":[4.4]}
])
alt.layer(
alt.Chart().mark_bar(color='#eee').encode(alt.X("ranges[2]:Q", scale=alt.Scale(nice=False), title=None)),
alt.Chart().mark_bar(color='#ddd').encode(x="ranges[1]:Q"),
alt.Chart().mark_bar(color='#bbb').encode(x="ranges[0]:Q"),
alt.Chart().mark_bar(color='steelblue', size=10).encode(x='measures[0]:Q'),
alt.Chart().mark_tick(color='black', size=12).encode(x='markers[0]:Q'),
data=df
).facet(
row=alt.Row("title:O", title='')
).resolve_scale(
x='independent'
)
